Design

This is a programming project that involves the implementation of a cloud computing program using Hadoop. In particular, I implemented a biomedical image analysis system to detect cells in a large number of images extract from an image list. The system utilizes the previously developed algorithm in Matlab as well as embraces the powerful cloud computing framework in Hadoop MapReduce. Even though the runtime has not been improved due to cluster configuration, the system has successfullly run cell detection using Hadoop. I have tested the system in a single-node-machine, and the results are satisfactory for detection purpose. Thoughout this project, I have learned many things about Hadoop API and its applications toward my research.
Data
- Over 200 data sequences.
- Each sequence contain at least 200 image frames.
- Each frame is a 1000 X 1000 pixel digital image.
- Total of possible processing data: 32 GB.
The figure shows a sample image frame.
Design Decisions
- To overcome programming languages incompatible, Matlab Code must be converted to .exe
- Map step will run detetion operation and likely to take most of running time.
- Reduce step will be executed as a identity reducer.
- Ideally, images in the same video sequence will be merged into a SequenceFile format that can be partition by Hadoop framework.
- Detection result is output as one .txt file.
Design Module
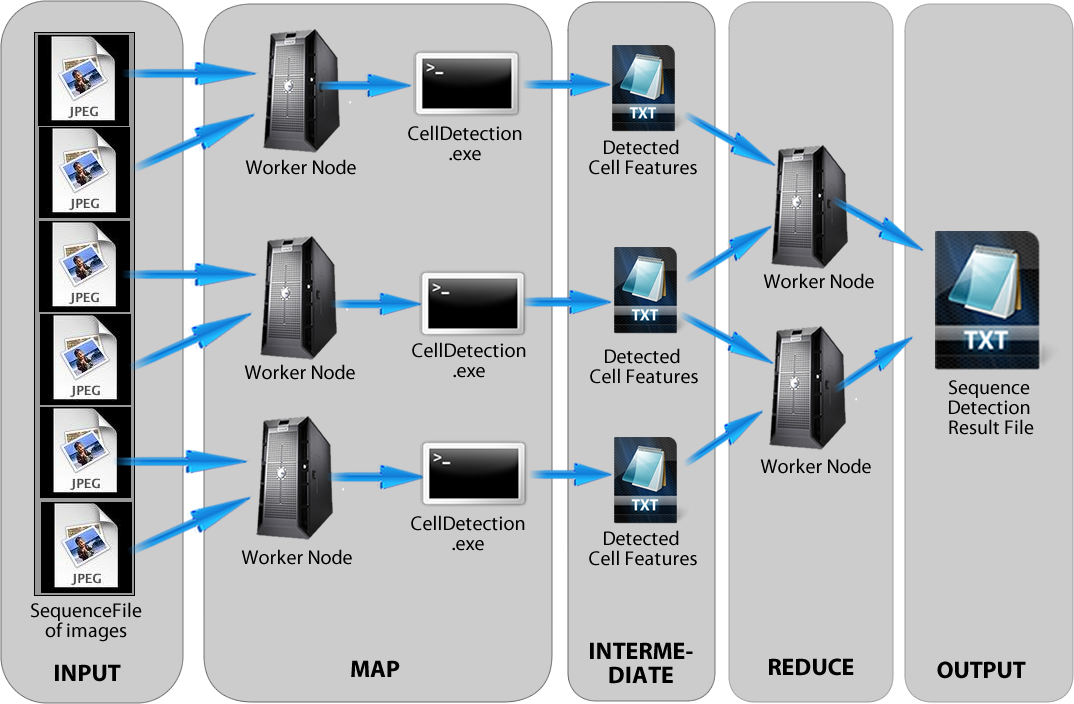
Implementation
Limitations
- Configured as one-node-cluster.
- Assumed the application knows where the image files located.
- Parallelization maynot be optimized until is operated in multiple-node cluster.
- Input as ImageName TextFile insteads of InputSequenceFileFormat.
Download
Project Code
Input File
Output File
Instruction for running
- $ javac -classpath Hadoop-0.19.0-core.jar -d rich CellDetection.java
- $ jar -cvf celldetection.jar -C rich .
- $ bin/hadoop jar celldetection.jar org.myorg.CellDetection input output
Results
Execution Screen Shot
Output Explanations
- Each image detection result is presented as a line in the output file.
- Output line format: ImageName, Cell 01 Info ~@~ Cell 02 Info ~@~ Cell 03 Info ~@~ ...
- Cell Info contains: cell ID, x-coordinate, y-coordinate, size, off-plane angle, long axis, and short axis
Visualization
Cell information results from the output is visualized in an input image. RED indicates cell detection by appreance GREEN indicates cell detection by motion
Q & A
- Q: What have I learned? A: There're several things that I have learned over the project timeline. First is to compile Matlab functions into executible file that can be run cross flatforms. Second is to call .exe file as a process within a Java program. Third, Hadoop MapReduce can be used as a simple parallelization process. And last but not least, searching the Java/ Hadoop API documentation can be very useful.
- Q: What's the most challenging obstacle ? A: I had a hard time configuring servers as well as set up input data the clusters HDFS. Perhalf it was the most challenging part for me. I still not figure it out due to lack of experience in server/network configuration.
- Q: Is there any interesting results that I found ? A: The matlab exectable run seamlessly as a process in a java application. I had implemented my detection algorithm,and was happy to see it's working. Although the MapReduce has not improve the performance significantly, but hopefully it will when the server configuration is done.
- Q: Where are useful resources that I discover while working on this project ? A: Cloudera video training is probably the most useful to learn about MapReduce Other resources are Hadoop Wiki and Hadoop 0.19.0 API
- Q: Why is this project significant ? A: Potentially, this project can be used to run our biomedical image data on the BioInformatics servers (upto 1000 nodes). Since I will be working with other collaborator from other discipline such as biologist and physician, having quicker way to analyze large amount of data make our research go forward.
- Q: How much work did I put in ? A: Over the timeline of the project which is approximately one month, I put work in converting matlab code to executible, calling .exe file from java, collecting detection features using Map step, attempting the configure the clusters, and making the report. As much as I wanted to put more work in, heavy courseload and research schedule has somewhat limited my availability to the project.
- Q: Why did the implementation not completely follow the design ? A: I learned to be more flexible with the implementation when the design is not feasible to be carried out in a time constraints. As software constantly need to be revolved, flexibility can be a good practice.
- Q: What to be improved in the future ? A: Several improvements can be done in the future. Fist thing is to configure the clusters so that this program can be run on multiple-nodes. Secondly, change the input format from list of images into SequenceFileFormat class.

